
BioHPC Cloud
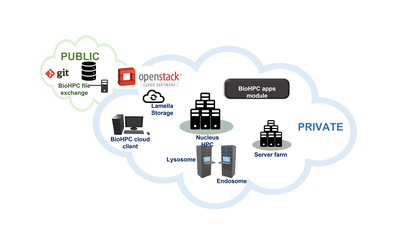
Compared to traditional HPC facilities, BioHPC aims to be easy-to-use for all of our users regardless of computing experience. Though the heart of our systems is a high-performance compute cluster with fast storage, we offer a wide range of services. The BioHPC Cloud encompases all of our systems and services, with access to:
- Simple web-based tools via the BioHPC Portal.
- Specialized, research-focused web applications, e.g. Astrocyte.
- Thin-client and workstation computers that are directly connected to the Nucleus compute cluster.
- Virtualized servers providing access to a multitude of research software.
- Traditional remote access using a command-line terminal and file transfer clients.
Nucleus Compute Cluster
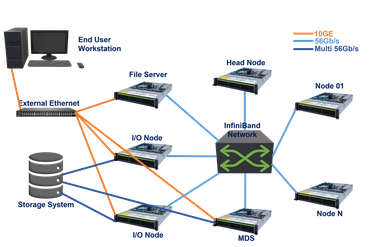
Our main compute facility is nucleus, a 800 node heterogenous compute cluster, consisting of:
| Partition | Nodes | Physical (Logical) Cores | Memory Capacity (GB) | GPU |
| 32GB | 280 | 16 (32) | 32 | N/A |
| 128GB | 24 | 16 (32) | 128 | N/A |
| 256GB | 78 | 24 (48) | 256 | N/A |
| 256GBv1 | 48 | 28 (56) | 256 | N/A |
| 384GB | 18 | 16 (32) | 384 | N/A |
| 512GB | 100 | 36 (72) | 512 | N/A |
| GPU | 40 | various | various | Tesla K20/K40 |
| GPUp4 | 16 | 36 (72) | 384 | Tesla P4 |
| GPUp40 | 16 | 36 (72) | 384 | Tesla P40 |
| GPUp100 | 12 | 28 (56) | 256 | Tesla P100 (2X) |
| GPUv100 | 2 | 36(72) | 384 | Tesla V100 16GB (2x) |
| GPUv100s | 10 | 36 (72) | 384 | Tesla V100 32GB (1x) |
| GPU4v100 | 12 | 36(72) | 376 | Tesla V100 32GB (4x) |
| GPUA100 | 16 | 36(72) | 1488 | Tesla A100 40GB (1x) |
| GPU4A100 | 10 | 36(72) | 1028 | Tesla A100 80GB (4x) |
| PHG | 8 | 24 (48) | 256 | N/A |
| Super | 432 | various | various | N/A |
Across these systems a total of >28,000 CPU cores and 100 GPU nodes are available.
Access to the cluster can be made through the Portal by an SSH login to the head node, nucleus.biohpc.swmed.edu, with a BioHPC username and password.
Lysosome Storage System
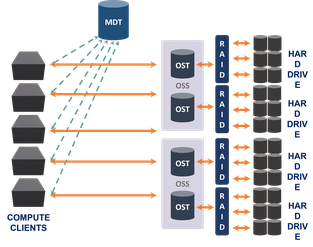
Lysosome is our primary high-performance bulk storage consisting of 5.4 PB raw storage provided by a Data Direct Networks SFA12X system, and an additional 960TB of raw storage provisioned with Dell PowerVault RAID hardware.
Our Data Direct Networks SFA12K system uses dual active-active storage controllers, connected to multiple storage enclosures. Providing raw speeds of 80-90GB/s the storage is configured to host a Lustre parallel filesystem and is connected to the nucleus cluster with multiple inifiniband and 10Gb ethernet links. 82 Object Storage Targets (backed by the DDN storage) are aggregated into a single high performance filesystem, with operations directed by a Meta Data Target (MDT). The architecture is well suited to typical workloads in BioHPC, which operate on large image and sequence datasets.
Endosome Storage System

Endosome is our off-site high-performance bulk storage consisting of 14 PB raw storage provided by IBM Elastic Storage Solution. Endosome uses IBM GPFS file system and locates at the QTS site at Irving, TX. This storage system is connected to our Nucleus cluster and Lysosome storage using high speed fibres across. Currently, it provides the /work and /archive file spaces for users and off-site backups of /home2 mirrors, /project incrementals and other web services for DR. This storage system has >1000 14 TB hard drives in 12 enclosures providing ~10 PB usable space. The actual aggregated IO throughput is ~60 GB/s.
Lamella Cloud Storage Gateway
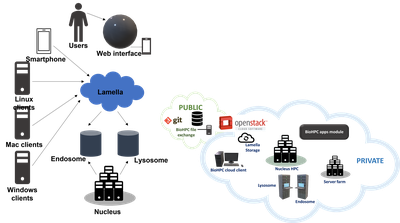
Lamella is our cloud storage gateway, providing easy access to files stored on BioHPC from Windows, Mac or Linux clients, over the web or on a smartphone. The Lamella web interface lamella.biohpc.swmed.edu at provides an easy-to-use site, similar to dropbox, to upload, download, manage and share files. Each user has an allocation of cloud storage and can also directly access their home, project, and work allocations. This service also supports file synchronization between computers using the OwnCloud client.
Direct access to files in home, project, and work locations is possible by mounting shared drives under Windows, Mac OS X, or Linux. Lamella shares these directories using SMB, with transfer rates up to 100MB/s for users on the campus 1 Gbps network and higher speeds available via the campus 10 Gbps network. FTP access to storage is also available via the Lamella gateway.
BioHPC Cloud Clients

BioHPC Cloud Clients are computers running Linux, integrated into the BioHPC cloud. These systems provide a graphical Linux desktop and have direct access to BioHPC storage over the campus 10 Gbps network. All software available on the BioHPC cluster can be used on the cloud clients, which are ideal for developing and testing code and analysis workflows. Scripts and programs can be run locally on the client or directly submitted to the nucleus cluster using the command. Access to Windows applications is possible with virtualization.
Two models of cloud client are available. The thin-client is a small desktop device, while the workstation is a larger tower PC. Currently, we have >70 BioHPC Cloud Clients campuswide. The workstation has a more powerful CPU than the thin-client and has an internal GPU card. The workstation is recommended for users running local analyses regularly or developing GPU code.
IB Network
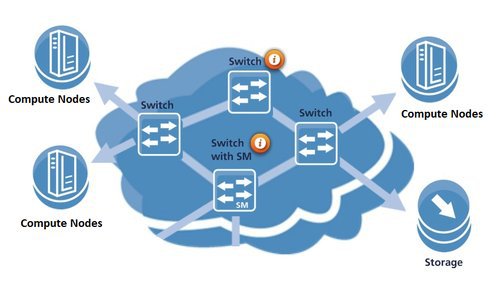
BioHPC Compute Nodes are interconnected via an Infiniband network in a fat-tree topology. The 10 HDR central swithes provide up to 80 Tbps total bandwidth. We are using HDR, EDR, FDR cables between 25 Mellanox switches and nodes and fiber cables between switches. The supporting port speeds can reach 56 Gbps and 100 Gbps respectively.