
Using the UCSC Genome Browser @ BioHPC
The UCSC Genome Browser is probably the most widely used public tool for browsing NGS data in a graphical format. The public web service at https://genome.ucsc.edu/ includes a wide range of public data, and the ability for users to display their own custom tracks. Custom tracks are used with the Genome Browser by making them availiable through an FTP or HTTP link, and then pasting this link into the custom tracks page. At UTSW there is a strict firewall so it has historically been difficult for groups to make their data available in a way that allows visualization on the UCSC public Genome Browser.
Following upgrades in early 2017 the lamella and cloud services can now serve files in a way that is compatible with the UCSC genome browser software. BioHPC users now have two options for working with custom tracks:
- Share a custom track file using a link in the lamella web interface, and visualize on the internal BioHPC UCSC Genome Browser mirror at http://genome.biohpc.swmed.edu
- Upload data to BioHPC's external file exchange site, share it with a link in the cloud.biohpc.swmed.edu web interface, and visualize on the public UCSC Genome Browser site. This method is not suitable for private or sensitive data.
Custom Tracks with BioHPC's UCSC Genome Browser Mirror
BioHPC hosts a UCSC Genome Browser mirror inside the university network at http://genome.biohpc.swmed.edu. This looks and works exactly like the main public site. The most common tracks are kept locally for speed. Less common tracks and genomes are retrieved as required. It is only accessible from the university network or via VPN and is appropriate for working with private or sensitive data that should not be exposed to 3rd parties (such as the public Genome Browser).
Let's go ahead and visualize a custom track using the BioHPC mirror. In this case it's a conservation wiggle file from UCSC.
Prepare the file to be shared from the lamella web interface
Make sure that the file you want to visualize in the browser is somewhere in your BioHPC storage. We recommend using /project or /work space as your /home2 directory has limited size, so it's not suitable for keeping large NGS files. In this example I copied the file to my project space, so it is located at:
/project/mydept/mylab/myuser/chr1.phastCons46way.placental.wigFix
Create a shared link for the file in the lamella web interface
Now login to the lamella web interface at https://lamella.biohpc.swmed.edu with your BioHPC username and password. If you have not already mounted your project or work space in the lamella interface please follow the instructions in the Introduction to BioHPC Guide..
In the main window of the web interface browse through your folders to find the file you want to visualize as a custom track:
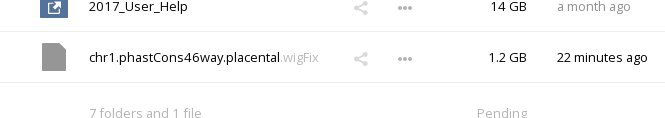
Click in a blank area next to the filename so that the sidebar opens up, and choose the 'Sharing' tab in the sidebar:
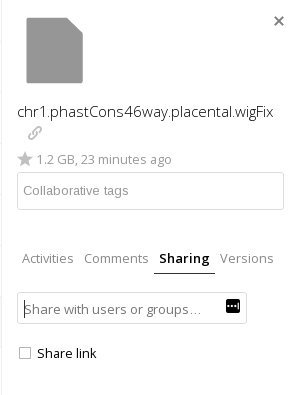
If you see a message that states 'Resharing is not allowed' make sure you checked the 'Allow Sharing' checkbox when you mounted your project or work space in the lamella web interface. Refer to the Introduction to BioHPC or Lamella Cloud Storage guides.
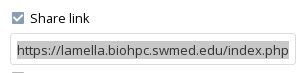
Now click the 'share link' checkbox. Copy the link that appears underneath it - this is needed to give the Genome Browser access to the custom track file. You may want to paste it into a text file stored safely somewhere for reference.
Add custom track on the UCSC Genome Browser Mirror
Now go to the BioHPC Genome Browser mirror at http://genome.biohpc.swmed.edu in your web browser. Choose the 'My Data -> Custom Tracks' option in the top menu bar:
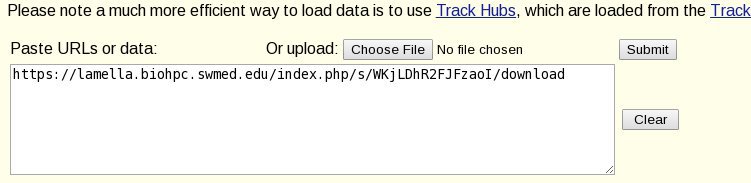
Paste the link from the lamella website into the 'Paste URLS or data' box on the page. Now add /download to the end of the link. This is important - the browser can't use the plain link as it directs to a sharing web page, not the file itself.
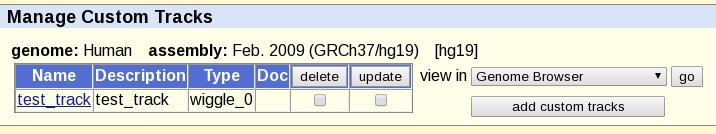
Click the 'submit' button and the mirror site will check that it can use the custom track file, and you should see the track in a custom track list:
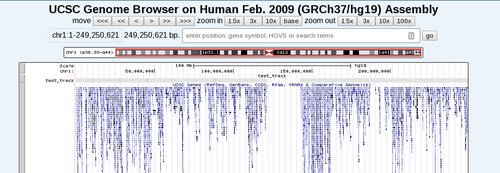
If all is well you can now proceed to the genome browser and open your region of interest. In this case we opened Chr1 (since the custom track only holds conservation data for this chromosome). The newly added custom track is displayed by default, and you can change the display format using the track options area below the main browser.
Custom Tracks with the Public UCSC Genome Browser
If the BioHPC internal Genome Browser mirror is not suitable for your work it is possible to visualize data on the public site https://genome.ucsc.edu/ by sharing your custom track from the external file exchange service at https://cloud.biohpc.swmed.edu
NOTE: By sharing data from cloud.biohpc.swmed.edu to the UCSC Genome Browser you are exposing it to an external server on the internet. You should not do this with sensitive or proprietary data - use the internal mirror instead.
To share to the public genome browser first login to cloud.biohpc.swmed.edu and upload your custom track files. Note that by default there is a 50GB quota on this system. When files have been uploaded to the file exchange service you may follow the instructions for the mirror above, but you must share the link from cloud.biohpc.swmed.edu and add the custom track on the public UCSC site instead of the BioHPC mirror.